Peering inside the black box by learning the relevance of many-body functions in neural network potentials
Abstract
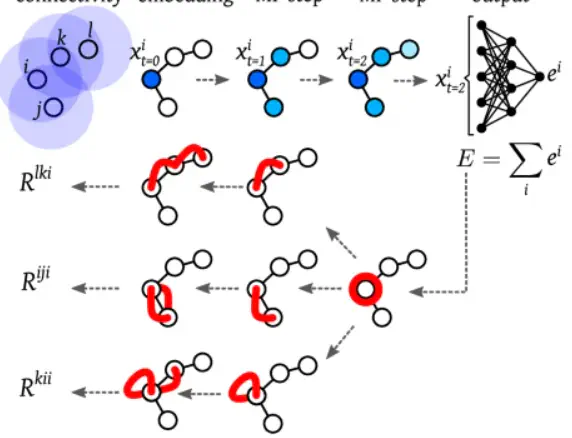
Machine learned potentials based on artificial neural networks are becoming a popular tool to define an effective energy model for complex systems, either incorporating electronic structure effects at the atomistic resolution, or effectively renormalizing part of the atomistic degrees of freedom at a coarse-grained resolution. One main criticism regarding neural network potentials is that their inferred energy is less interpretable than in traditional approaches, which use simpler and more transparent functional forms. Here we address this problem by extending tools recently proposed in the nascent field of explainable artificial intelligence to coarse-grained potentials based on graph neural networks. With these tools, neural network potentials can be practically decomposed into n-body interactions, providing a human understandable interpretation without compromising predictive power. We demonstrate the approach on three different coarse-grained systems including two fluids (methane and water) and the protein NTL9. The obtained interpretations suggest that well-trained neural network potentials learn physical interactions, which are consistent with fundamental principles.